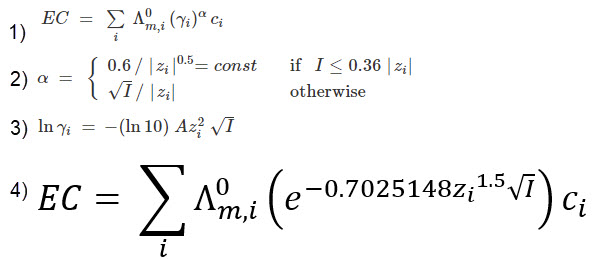Electrolyte Conductivity vs. Ionic Activity: Why EC Alone Can Mislead Your Nutrient Decisions
Your EC meter is telling you only part of the story. Two nutrient solutions reading identical EC values can produce dramatically different plant growth outcomes in controlled studies. The reason lies in a fundamental measurement limitation: electrical conductivity reports total dissolved ions without distinguishing nutrient species from growth-limiting salts. This bulk measurement masks the specific ionic composition that drives membrane transport, competitive inhibition at root uptake sites, and toxicity thresholds. Understanding what EC actually measures will help you recognize when additional monitoring becomes necessary.

EC measures bulk conductivity, not what plants actually absorb
Electrical conductivity provides an indiscriminate measure of total dissolved ions in solution. Your meter detects all charged particles without distinguishing whether they are essential nutrients or growth-limiting salts. As detailed in a review on ion-selective sensing in controlled environment agriculture, EC cannot differentiate among nutrient species, and different ions contribute disproportionately to measured values (1).
Why EC alone proves insufficient has multiple explanations. Ion identity matters: sodium and chloride at high concentrations cause specific toxicities independent of osmotic effects. Ion ratios matter: excess potassium competitively inhibits calcium and magnesium uptake at membrane transporters. And the effective concentration of ions in solution, termed ionic activity, also plays a role. Activity represents the concentration available for chemical reactions, always lower than measured concentration due to ionic interactions in solution.
Plants do not directly sense ionic activity. They respond to membrane transport kinetics, electrochemical gradients, competitive inhibition at transporters, and rhizosphere chemistry. Ionic activity influences these processes, but ion identity, ratios, and specific toxicities provide the more actionable framework for understanding when EC measurements mislead.
| Parameter | What It Measures | Plant Relevance |
|---|---|---|
| EC (electrical conductivity) | Total dissolved ion charge carriers | Indirect indicator only |
| Ion concentration | Absolute quantity of each ion species | Laboratory reference value |
| Ionic activity | Effective concentration for chemical reactions | Influences uptake kinetics and ion availability |
The Debye-Hückel equation predicts activity coefficient changes with ionic strength in ideal solutions (1). At typical nutrient solution concentrations, divalent cations like calcium and magnesium might show activity coefficients around 0.36, suggesting reduced effective availability.
However, Debye-Hückel works best at low ionic strength with simple solutions. Real hydroponic systems are multi-ion mixtures with chelators, buffers, and temperature fluctuations. Activity coefficients are not static, generalizable values. The conceptual value is recognizing that concentrated solutions have reduced effective nutrient concentrations, with divalent ions more affected than monovalent ones. But this thermodynamic consideration is only part of why EC measurements can mislead. Ion-specific toxicities, competitive uptake, and ratio imbalances often matter more in practice.
Identical EC readings can mask specific ion toxicities
The clearest evidence that EC measurements conceal important information comes from controlled salt stress experiments comparing solutions matched for EC but differing in ionic composition. Research on faba bean exposed plants to sodium-dominant, chloride-dominant, and sodium chloride treatments, all maintained at the same EC range of 8.4 to 9.0 dS/m with identical osmotic potentials (2).
These were deliberately extreme compositions designed to test toxicity mechanisms, not optimized fertigation protocols. The results show what EC masks under stress conditions. At matched EC levels, chloride-dominant solutions reduced shoot dry weight by 24 to 40 percent compared to controls, while sodium-dominant solutions caused only 5 to 23 percent reduction. The NaCl treatment combining both ions produced the largest growth inhibition at 36 to 55 percent, demonstrating additive toxicity effects (2).
| Salt Composition | EC (dS/m) | Osmotic Potential (MPa) | Shoot Dry Weight Reduction |
|---|---|---|---|
| Sodium-dominant (Na₂SO₄, Na₂HPO₄, NaNO₃) | 8.8 | -0.49 | 5-23% |
| Chloride-dominant (CaCl₂, MgCl₂, KCl) | 8.4 | -0.48 | 24-40% |
| NaCl combined | 9.0 | -0.50 | 36-55% |
The point is not that growers routinely leave 40% yield on the table by relying on EC. The point is that EC provides no information about which specific ions contribute to the measured value. Two solutions at identical EC can have completely different ionic compositions, and those differences matter when toxic ions accumulate or when antagonistic interactions suppress nutrient uptake. The experiments demonstrate that specific ion toxicity operates independently of bulk conductivity measurements.
Activity coefficients and competitive uptake
Plant nutrient uptake follows Michaelis-Menten kinetics, with roots responding to effective ionic concentrations at membrane transport sites. Research on ion uptake kinetics across crop species found that uptake rates depend on transporter properties and the concentration gradients driving diffusion and active transport (3).
However, plants are not passive. They actively regulate transporter expression in response to nutrient status. Root exudates, rhizosphere pH shifts, and microbial interactions create a dynamic environment that activity coefficients alone cannot predict. In recirculating systems, root-zone biology often dominates availability more than solution thermodynamics.
Each nutrient ion has an optimal concentration range. Deviation causes deficiency or toxicity. High potassium suppresses magnesium and calcium uptake through competitive inhibition at transporters, even when those nutrients appear adequate (1). This operates through membrane competition rather than activity coefficients.
The charge on an ion affects both its activity coefficient and its behavior at root membranes:
| Ion Charge | Example Ions | Activity Coefficient at I = 0.01 M | Activity Coefficient at I = 0.1 M |
|---|---|---|---|
| Monovalent (+1) | K⁺, NO₃⁻, Na⁺ | ~0.90 | ~0.76 |
| Divalent (+2) | Ca²⁺, Mg²⁺, SO₄²⁻ | ~0.68 | ~0.36 |
| Trivalent (+3) | Fe³⁺, Al³⁺ | ~0.45 | ~0.04 |
Calcium and magnesium deficiencies can appear in high-EC systems even when solution analysis shows adequate concentrations. Multiple factors contribute: reduced activity coefficients at elevated ionic strength, competitive inhibition from excess monovalent cations, precipitation reducing free ions, and inadequate transporter expression in some cases.
A practical framework for knowing when EC suffices
Understanding EC limitations does not mean abandoning it as a management tool. The question is when EC monitoring alone provides adequate control and when additional measurements become necessary.
EC works adequately when:
- Using stable, tested nutrient recipes with known water sources
- Operating within established EC ranges for your crop (typically 1.5-2.5 dS/m for most vegetables)
- Observing normal growth with no unexplained deficiency or toxicity symptoms
- Running drain-to-waste systems where solution composition stays close to input values
Move beyond EC-only monitoring when:
- Source water contains significant sodium, chloride, or bicarbonate (>50 ppm of concerning ions)
- Running recirculating systems where selective uptake changes ratios over time
- Pushing high EC strategies (>3.0 dS/m) for crop steering or stress conditioning
- Observing nutrient disorders that do not resolve with EC adjustments
- Using fertilizer blends high in chloride-based salts (muriate of potash, calcium chloride)
Monitor ion ratios alongside EC. Track potassium to calcium ratios (typically 1:0.7 to 1:1 molar basis for greenhouse vegetables), calcium to magnesium around 3:1 to 5:1, and watch for sodium and chloride accumulation. These targets vary by crop, growth stage, temperature, and transpiration rates, but maintaining balanced ratios matters for preventing competitive uptake regardless of activity calculations.
Account for ionic strength effects on divalent nutrients. When operating at elevated EC for generative strategies, calcium and magnesium may require 10-20% higher concentrations above 2.5 dS/m.
Consider periodic solution analysis. Laboratory testing provides ground truth for whether EC correlates with intended composition. Test quarterly for established protocols, monthly when developing new strategies (1).
Watch for ion-specific symptoms. Chloride toxicity produces marginal leaf burn, sodium affects older leaves first, calcium deficiency appears in growing points. When symptoms appear at moderate EC with no disease, investigate ionic composition.
The measurement matters, but so does the biology
The hydroponic industry invested heavily in EC monitoring because it is simple and inexpensive. This created reliance on a parameter that cannot distinguish nutrient species from non-nutrient salts. Plant roots respond to individual ions through specific transporters, adjust those transporters based on status, and modify rhizosphere chemistry (3).
Understanding ionic activity provides one lens for recognizing EC limitations, but ion identity, ratios, and toxicities matter more for practical management. The primary insight is simpler: EC cannot tell you which ions are present or whether problematic species like sodium and chloride are accumulating.
The practical approach combines EC monitoring with awareness of when it suffices. For stable systems with proven recipes and clean water, EC provides adequate control. When water quality varies, in recirculating systems with selective depletion, or when pushing high-EC strategies, monitor individual ions. Two growers at identical EC will achieve different results based on water quality, fertilizer choices, and ionic composition.
Research on matched-EC salt stress shows specific ion toxicities operate independently of bulk conductivity. Your EC meter remains useful for routine monitoring, but recognizing its limits prevents misdiagnosis. Understanding that EC measures total ions rather than ion identity or ratios transforms it from a complete system into one point within a fuller framework.